Background and Introduction
Ramsey Theory is the study of combinatorial problems proposed by Frank Ramsey in 1930. The version of his problem as applied to graphs asks, “what is the smallest complete graph such that there is at least one clique of a given size and color?” This can be represented by where x and y are the sizes of the cliques to find for red and blue, respectively.
means to find the smallest graph such that we are guaranteed either a red 3-clique (3 vertices connected together at every point, or a complete graph on 3 vertices) or a blue 3-clique. This has been shown to be 6. We can show this by providing an example that a complete graph on 5 vertices (
) can be colored such that it contains no red or blue 3-cliques. Then, we show that for every coloring of
, there must necessarily be a red triangle or a blue triangle in the graph. This can be verified computationally or combinatorially. (See the nice example here: Theorem on friends and strangers)
While the problem is relatively easy to solve for a small value like , the complexity of the problem increases greatly when considering larger clique sizes. For instance,
is 18, and we only know that the value of
is somewhere between 43 and 49 inclusive. Because the number of cases one must check increases exponentially with each increase in clique size, it becomes impractical for traditional computing very quickly.
Therefore, we will utilize a genetic algorithm in an attempt to verify and perhaps improve the lower bound of . To do this, the program must complete the following:
-
Use the genetic algorithm to test for graphs that have low numbers of cliques
-
Perform exhaustive testing on these graphs, and if we can find even one coloring where a clique does not exist, we will have shown that
.
Implementation
My algorithm is implemented using Java. Graphs are represented using an adjacency matrix, and a coloring of the graph (ColorMatrix) are 2D boolean arrays. In this scheme, coloring[x][y]==0 represents a blue edge from x to y and coloring[x][y]==1 represents a red edge. The colorings are symmetric; that is, coloring[x][y] == coloring[y][x]. These colorings form a base for my Chromosome class, which is used in the genetic algorithm to represent a “piece” of information that can be mated with other chromosomes, mutated, and scored based on the number of cliques found. A population stores multiple chromosomes.
My basic algorithm works as follows:
- Create a population of random colorings of
-
Check a set of edges for 5-cliques (how these sets are determined will be discussed later). Assign a fitness score based on the number of cliques found
-
Crossover/mutate the chromosomes
-
Run this many times until every member of the population has fitness 0 (no cliques found in that set of edges)
-
Take these “possible zeroes” and begin to test more edges to find cliques
-
if we find any new cliques, this coloring is no longer any good to us (we are looking ultimately for a coloring with no cliques whatsoever)
-
pare down the population to a set of graphs that have no cliques in set 1 and set 2
-
eventually this leads to an exhaustive search but by that point there will be only a few “likely” candidates that passed every test set (this saves computation time rather than checking every clique right off the bat)
-
Calculating Fitness
The fitness function for our genetic algorithm checks a set of edges in the graph to see if there are any cliques. We test from “test sets” instead of checking every clique every time. The reasoning behind this is that if a clique is found on the first test, we need not go any farther. The multiple-stage fitness testing allows us to prune out “bad” data and get more likely solutions to our problem. The first test set data is computed as follows (each 5-tuple represents a set of five edges to check for a clique — that is, all the same color)
We use a simple method of counting up iteratively the first two items in the list, then using increasing consecutive numbers for the rest of the clique. This way, lots of cliques are tested, but this test set leaves out cliques that are not formed from consecutive edges (e.g., would not be tested in this list). Once the list of edges to check is generated, we simply test the edges on each node in the set. If the colors are all the same, then we have a clique and add to our fitness.
The second test set would be applied after the population has converged for the first time to zeroes, and this time counts up by multiples of 2 (so would be a viable choice in this set).
An alternative fitness function was considered that involves inserting vertices into a binary search tree based on their coloring values. To do this, first one creates a random permutation of edges (values 0-42). Then using the colorings (0 and 1 instead of positive and negative) we can insert the nodes either left or right in a binary search tree with the first node in the list as the root. By looking at the output of this tree, we can get a visual representation of color patterns. The idea is that by finding a long “path” in one of these trees we know that it has seen the same color many times, thus suggesting a clique exists.
In the below example, the set forms a long “path”, meaning that each edge is the same color.
Unfortunately, this implementation is not currently functional in my program.
Parent Selection and Crossover
The backbone of any genetic algorithm is the crossover. This models the mating process in traditional biology and is used to make the Chromosomes (solutions) improve themselves over many generations by keeping successful traits and discarding others. Parents are selected in my model randomly from the population, but there is a small chance (about 5%) that the best member of the population would be chosen as a parent instead. This provides a slight bias toward better traits but still allows for plenty of randomness and new data.
The crossover is a simple one. Using two Chromosomes (colorings of the graph) we use a random value for each x, y in the colorMatrix to determine whether the “mom” or the “pop” will contribute their genes (coloring[x][y]). There is a 50-50 chance of inserting mom(x,y) into the baby as opposed to pop(x,y). The “baby” Chromosome results in a mixture of data from both parents, and if the new Chromosome’s fitness (number of cliques) is lower than the current worst value in the population, we replace that value with our new baby. This allows the population to improve over time.
Our mutation method simply flips a bit in the coloring based on a random value. The mutation rate was 0.08, or 8%.
Problems and Solutions
One of the things I quickly noticed when building this project was that the fitness function, although it does not brute force every possible clique check, can still take a prohibitively long time at higher population sizes. The getFitness() function is called a bit too liberally, and saving the fitness value to retrieve later would improve the efficiency in this area.
Another downfall is that since we are only testing a limited set of cliques each time, it is possible (although desirable, given the current approach) to “learn the data”, and evolve a solution that contains no cliques in the tested area, but may have cliques elsewhere. While this seems like a problem at first glance, we can use this to our advantage because having the algorithm “learn” each test data set is what allows us to prune the search space and find possible colorings that do not contain cliques. If we decided this was an undesirable behavior however, we could change the fitness function to include x random permutations of edges each time. The problem with this method is that we could never be sure that a clique certainly did not exist in a certain area over multiple rounds, as the sets tested would be randomized each time. Finally, it is possible to go with the old-fashioned approach and simply check every clique after all.
Future Work
While this program currently provides a proof of concept and implementation up to the first round of test data, there is much room for expanding this project. First of all, multiple rounds need to be completed within the algorithm, so that we can generate colorings that are more likely to have no cliques. Other things that would be useful are outputting data to a text file for further reading/analysis, and performing general optimizations throughout my code for performance.
In addition, I would really like to get a working implementation of the Alternate Fitness function implemented and working. I feel like this is an interesting direction to take the project and it would be helpful to have a more visual representation of where cliques are occurring.
While the population initialization currently creates a population of random colorings, it would be interesting to see what happens when we change the ratios of red edges to blue edges in the chromosomes, starting at initialization. What if each coloring was required to be within a certain amount red and a certain amount blue? This is definitely something to check into as well.
Conclusion
Currently, we are able to generate lists of colorings that do not have cliques within the first test set of edges. As we add on more edge sets to test, we will create more constrained lists. Further testing will reveal if any of these “possible” solutions truly do not contain such cliques. For this, an exhaustive search is the only way. Although it is on the order of 900,000 edge combinations () to check, this is time consuming but doable, especially if we have a relatively small set of graphs to check.
Finally, the code is available on https://github.com/nelsonam/ramsey if anyone is interested. Share your comments in the box below and if you have any questions feel free to ask!
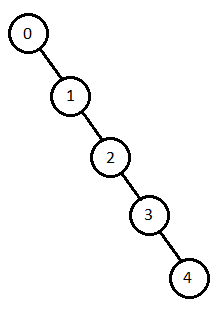
[…] response to my previous article about genetic algorithms for Ramsey theory, a few readers asked me to give a bit more of an introduction about genetic algorithms. As such, […]
[…] have seen some of these difficult problems before in previous posts: Genetic Algorithms for Ramsey Theory and The Travelling Santa Problem, as well as Introduction to Genetic Algorithms all have good […]
[…] my last post on this topic, I discussed how I was working on a genetic algorithm to search mathematical graphs […]